Greeting

Hisae Tateishi-Karimata
Frontier Institute for Biomolecular Engineering Research (FIBER),
Konan University
Until now, nucleic acids comprised in the genome have been considered molecular units that contain all genetic information. In contrast, proteins are believed to be to regulators of gene expression. However, in recent years, several reports have described non-canonical structures of nucleic acids (triplex, G-quadruplex, cruciform structures, and among others) that can regulate gene expression in human cells, which may overturn this preconceived notion. For example, the formation of G-quadruplexes in oncogenes can suppress its expression. The roles of nucleic acid structures as promoters of cancer is now attracting attention, and novel drugs targeting nucleic acid complex structures are being explored worldwide.
It is known that the structure of nucleic acids changes dynamically under the influence of the surrounding environment. We have considered that, in all living organisms, nucleic acids sense the surrounding environment, change their structure multidimensionally, and proactively regulate their expression. In previous research, the structure and function of nucleic acids have been analyzed for each species; however, from a physicochemical perspective, the mechanism of nucleic acid structure formation is considered to be species-independent. Nevertheless, to the best of our knowledge, no study to date has analyzed similarities and differences in gene expression mechanisms by non-canonical structures beyond the framework of biological species.
In this research area, we will conduct new investigations aiming at the following two points:
1) Elucidation of the "dimensional response genome”
We will focus on nucleic acid structures that fluctuate in a multidimensional manner in response to the environment and elucidate the molecular mechanisms that regulate gene expression from a physicochemical point of view, regardless of the biological species framework, which we call "dimensional response.” To this end, we aim to propose the existence and significance of a new genomic function based on nucleic acid structures, called “dimensional response genome," that are transversal to all disciplines that deal with life.
2)Development of a genome bank (DiR-GB) that can predict and use the dimensional response genome
We will explain the unified expression regulation mechanisms independent of species and construct a genome bank (Dimension Responsive Genome Bank [DiR-GB]) that will gather all the information.
The DiR-GB will deepen our understanding of the roles of nucleic acid structures in human cells and is expected to be used to predict multidimensional responses of targeted organisms (not only humans, but also viruses and plants) for development of technologies to control biological phenomena. The DiR-GB will be made public so it can be freely used by researchers worldwide in a wide range of fields, including medical engineering, agriculture, and materials chemistry.
Project Outline
In this research project, we will integrate research approaches from other fields and promote the following research in a stepwise manner (Figure 1).
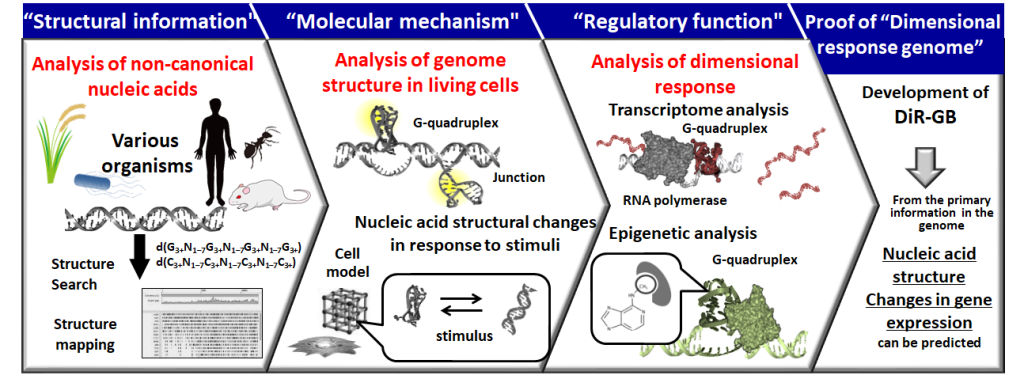
[1] Using analytical chemistry and information science approaches, we will analyze the information from various organisms whose whole genome sequences have been deciphered and which can form the non-canonical structure of nucleic acids. The "structural information" of nucleic acids that show multiple responses will be determined.
[2] Using physical chemistry, biochemistry, and inorganic materials science approaches, we will understand the "molecular mechanism" of nucleic acid structure-dependent dimensional responses based on direct observation of non-canonical structures in living cells and physicochemical parameters of nucleic acid structure changes in response to environmental changes in cellular model systems.
[3] Using molecular biology and phytochemistry approaches, we will analyze gene expression changes in response to non-canonical structures and correlate them with phenotypic changes in cells and individuals to understand the "regulatory function" of biological phenomena by multiple responses.
The findings will be integrated to create the Dimension Responsive Genome Bank (DiR-GB) — the first data bank of nucleic acid structures that show dimensional responses in various species.
A01 Group
Comprehensive analysis of nucleic acid structures involved in modulation of gene expression across species
In our group, A01, we aim to demonstrate that nucleic acid structures, which are responsible for modulating gene expression, are present in various species and are optimized to function in each living condition by conducting genome-wide and transcriptome-wide analyses of their gene expression. In particular, we will discuss functional similarities and differences within nucleic acid structures among species using bioinformatic approaches, which will represent the foundations for further physicochemical and biochemical analyses to be conducted by other groups.
-
Principal Investigator

Tamaki EndohAssociate ProfessorResearch Map
Frontier Institute for Biomolecular Engineering Research (FIBER), Konan University
Research Interest: Cellular engineering, Biomolecular engineeringRoles in this research area
Genome-wide and transcriptome-wide analyses of gene expression influenced by nucleic acid structures -
Co-Investigator

Yiwei LingAssistant ProfessorResearch Map
Medical AI center, Niigata University School of Medicine
Research Interest: BioinformaticsRoles in this research area
Bioinformatic analyses of nucleic acid structures involved in gene regulation -
Research Collaborator

Shujiro OkudaProfessorResearch Map
Medical AI center, Niigata University School of Medicine
Research Interest: BioinformaticsRoles in this research area
Bioinformatic analyses of nucleic acid structures involved in gene regulation
A02 Group
Elucidation of the mechanism for dimensional response genome using intracellular environmental evaluation systems
-
Principal Investigator
Hisae Tateishi-KarimataAssociate ProfessorResearch map
Frontier Institute for Biomolecular Engineering Research (FIBER), Konan University
Research Interest: Functional nucleic acid chemistryRoles in this research area
Elucidation of the underlying mechanism of the dimensional response genome using intracellular environmental evaluation systems -
Co-Investigator

Takaaki TsuruokaAssociate ProfessorResearch map
Frontiers of Innovative Research in Science and Technology (FIRST), Konan University
Research Interest: Inorganic materials scienceRoles in this research area
Evaluation of the behavior of nucleic acids in MOF microspace -
Research Collaborator

Saki MatsumotoAssistant ProfessorResearch map
Frontier Institute for Biomolecular Engineering Research (FIBER), Konan University
Research Interest: Nucleic acid chemistry, Organic chemistry
A03 Group
Elucidation of multidimensional physiological functions of nucleic acid structures
-
Principal Investigator

Miki ImanishiAssociate ProfessorResearch map
Institute for Chemical Research, Kyoto University
Research Interest: Epigenetic and epitranscriptomic regulation, Nucleic acid binding proteinsRoles in this research area
Elucidation of the intracellular functions of the non-canonical structures of nucleic acids -
Co-Investigator

Shiori S Aki Assistant Professor Research map
Graduate School of Science and Technology, Nara Institute of Science and Technology
Research Interest: Plant molecular biologyRoles in this research area
The functional analysis of the G4 structure in plants

